Part 3 - Adding the Details
The CBM Jmol Training Guide

Introduction
At this point, we have opened a protein structure, explored it using the display format and color commands, and then used the select commands to simplify the design and focus on the key regions and features that will help tell our molecular story.
But often there are details that when added back to a backbone model design will help emphasize the molecular story. In this section of the Jmol Training Guide, you will learn how to add key amino acid sidechains, substrates or other small molecules, and molecular bonds to your protein model design, as shown in the image below and to the right.
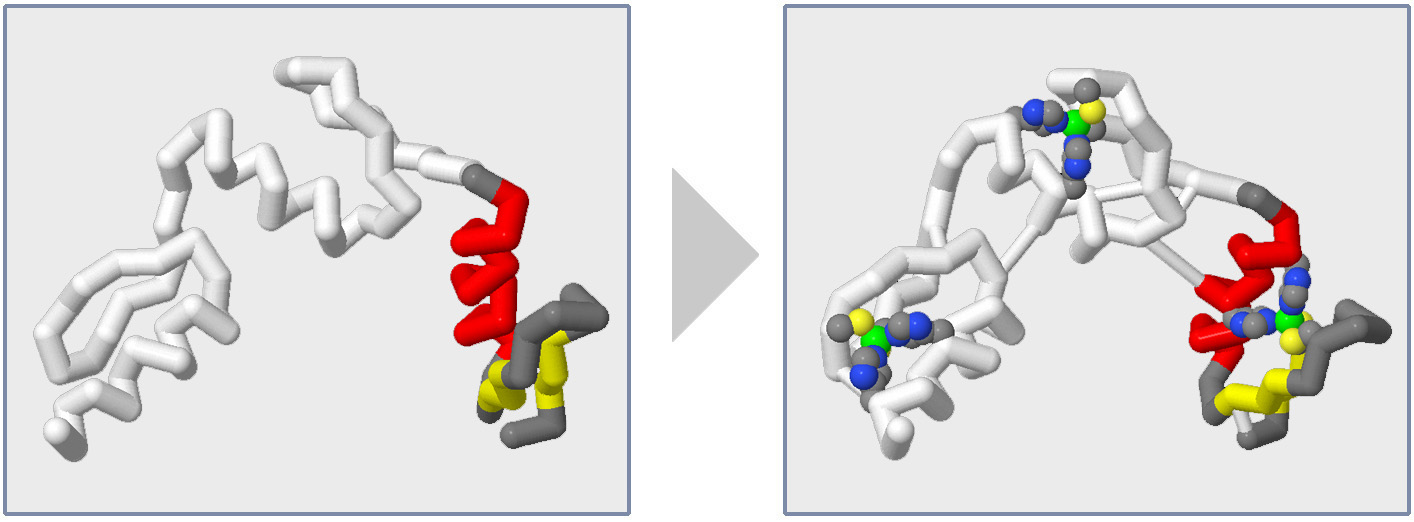
Adding Sidechains
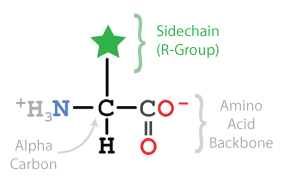
Proteins are long linear sequences of linked amino acids that fold up into complex 3-dimensional shapes based on the order of their amino acids. Because of this, the specific amino acids in a protein are very important and directly relate to the protein's structure (shape) and function (job).
Most protein models designed with the CBM's Jmol User Design Environment for 3D Printing use Backbone Format. In backbone format, only the position of the alpha carbon in each amino acid is shown, representing a bend or kink in the backbone. All of the other atoms within the amino acid are not displayed, including the Sidechain, also called the R-Group.
But sometimes you may wish to display a specific amino acid residue's sidechain because it is important to telling the story of your protein, such as a sidechain that binds to a substrate or holds the protein together, even if the rest of your protein structure remains represented in backbone format.

To select and display a specific sidechain, you will use the select command followed by the amino acid residue number and end with and sidechain. We recommend giving sidechains a spacefill of 1.25, which is large enough to hold them together and make them sturdy if 3D printed, but small enough to keep them from overwhelming the other features of your model design. We also recommend using color cpk to color your sidechain by atom type.
To add a sidechain for Histidine 29 in the 1zaa.pdb zinc finger protein:
select his29 and sidechain
spacefill 1.25
color cpk

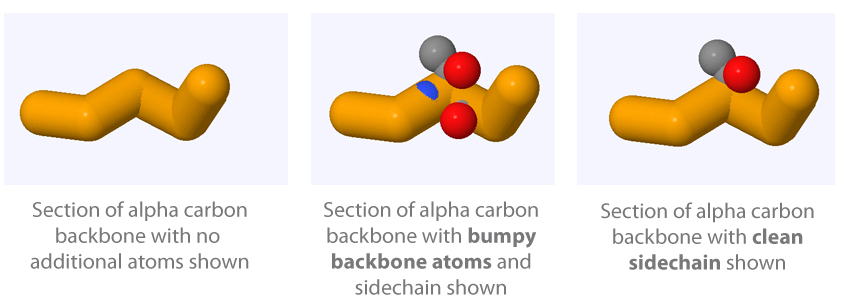
A "Bumpy" Backbone
If you were to add your sidechain without using the and sidechain syntax when selecting, it would produce a bumpy backbone. This means that the backbone atoms of the amino acid would be displayed, and partially show through the alpha carbon backbone display. This is aesthetically distracting, and therefore should be avoided unless there is a specific reason you would like to display the backbone atoms.
Adding Ligands or Substrates
Many proteins interact with small molecules that are not part of the protein chains. These small molecules are often included in molecular structure files and can be selected and displayed in Jmol. Every small molecule in a structure file is given a unique two or three-letter identifier that can be used to select them through the Jmol console.
For example, the 1zaa.pdb zinc finger structure has three zinc atoms, each with the two-letter identifier zn. To select and display these three zinc atoms:
select zn
spacefill 1.25
color green

There are two main ways to determine what the unique identifier is for any given small molecule in a structure:
-
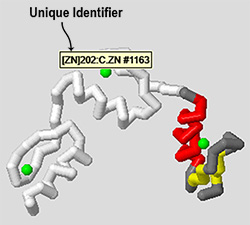
Just like any atom you see displayed in Jmol, you can roll your mouse cursor over the molecule and it will display a bit of information about the atoms your are hovering over. The very first piece of information will tell you the unique identifier to use when selecting that molecule in the Jmol console.
-
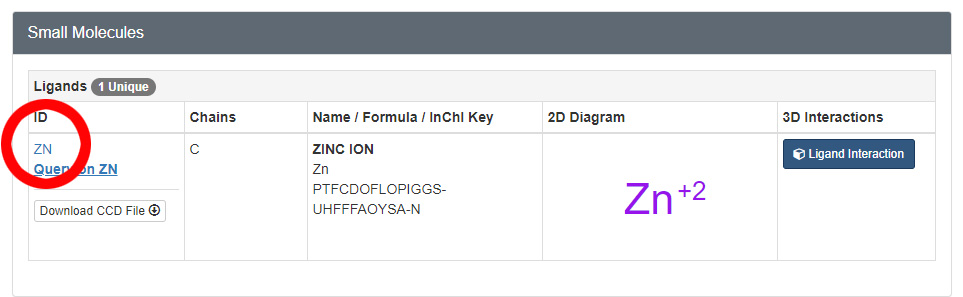
If you look at the RCSB Protein Databank's entry page for the structure file you are using, you will find a section about half way down the page titled Small Molecules. This section will list every small molecule in the structure file, including their unique identifiers. For example, the 1zaa.pdb zinc finger structure page lists the zinc atoms and the unique "zn" identifier (circled in red in the image below).
Adding Molecular Bonds
In addition to covalent bonds between atoms in a molecule, Jmol has the ability to render hydrogen bonds and disulfide bonds. These bonds can often help highlight the rigidity of secondary structures (specifically, beta pleated sheets) as well as highlight key structure features of certain proteins.

Hydrogen Bonds
To add hydrogen bonds to secondary structures within your model, you will use the calculate hbonds command, followed by a series of formatting commands that will make the hydrogen bonds solid and correctly position them on your protein backbone.
We suggest that you typically do not add hydrogen bonds to alpha helices, since they do not add stability to the model and actually clutter the view of the structure. Adding hydrogen bonds to beta sheets, however, provides additional support for the final model and is recommended. We also suggest using a subtle color like white or gray for your hydrogen bonds so they are not visually overwhelming, and recommend a size of 0.75 angstroms, which compliments a backbone of 1.25 angstroms.
select sheet
calculate hbonds
set hbonds solid
set hbonds backbone
hbonds 0.75
color hbonds white
Note: Jmol recognizes both hbonds and hbond (singular) when entering commands. Also note that all of the commands demonstrated above will only affect hydrogen bonds in the currently selected area of the molecular structure.

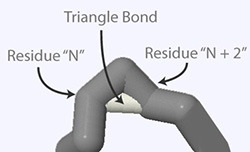
To remove hydrogen bonds use the hbonds off command, which will remove the hydrogen bonds in the currently selected area of the structure. To remove just a single hydrogen bond, select only the two amino acids that the hydrogen bond connects and then use the hbonds off command.
When Jmol calculates hydrogen bonds, it occasionally inserts a hydrogen bond between two amino acids on the same strand, with a single amino acid between the two. These types of hydrogen bonds create what looks like a triangle between two kinks of the alpha carbon backbone and are therefore referred to as triangle bonds. For aesthetic reasons, we suggest that triangle bonds be identified and removed from your structure.

Disulfide Bonds
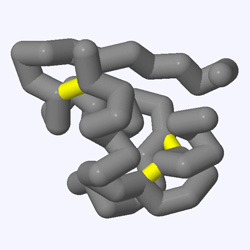
Some protein structures will have disulfide bonds, which form between two cysteine amino acids that lie close to each other. Disulfide bonds often stabilize the structure of small or thin proteins and are sometimes even involved in the protein's function.
The commands used to display disulfide bonds are very similar to the commands used to display and format hydrogen bonds, except with "ssbonds" instead of "hbonds", and you do not need the "calculate" or "set solid" commands.
select all
set ssbonds backbone
ssbonds 0.75
color ssbonds yellow

An Example Design
The entire goal of adding select sidechains, small molecules and molecular bonds to your design is to highlight key features that will help you clearly communicate a molecular story.
For example, we could revisit our 1zaa.pdb zinc finger design from the previous section of this Jmol Training Guide and add a few of the key sidechains that interact with the zinc atom, which are crucial to holding this small protein together. The commands below would produce a model that highlights these key features.
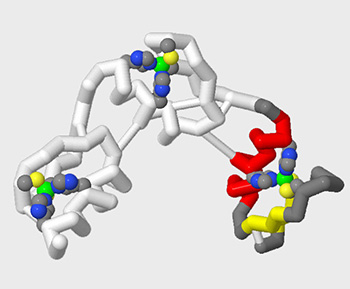
select all
backbone off
wireframe off
spacefill off
select :c
backbone 1.25
color white
select :c and 1-31
color gray
select helix and 1-31
color red
select sheet and 1-31
color yellow
select zn
spacefill 1.25
color green
select sheets
calculate hbonds
set hbonds solid
set hbond backbone
hbond 0.75
color hbond white
select his25 and sidechain
spacefill 1.25
select his29 and sidechain
spacefill 1.25
select his54 and sidechain
spacefill 1.25
select his57 and sidechain
spacefill 1.25
select his81 and sidechain
spacefill 1.25
select his85 and sidechain
spacefill 1.25
select cys7 and sidechain
spacefill 1.25
select cys12 and sidechain
spacefill 1.25
select cys37 and sidechain
spacefill 1.25
select cys40 and sidechain
spacefill 1.25
select cys65 and sidechain
spacefill 1.25
select cys68 and sidechain
spacefill 1.25
select sidechain
color cpk
Conclusion
At this point, you should be able to open molecular structures in Jmol, manipulate their display formats and colors, focus in on important regions using the select command, and add back key sidechains, small molecules and molecular bonds that help communicate your molecular story.
In the next section, 3D Printing Your Design, you will learn how to prepare your design for 3D printing, and either print the model yourself or order it from a 3D printing service.

Table of Contents
