The 2020-21 Theme:
CRISPR and Base Editing
 Be extra sure to explore the information in all three tabs below
Be extra sure to explore the information in all three tabs below
Welcome!
To the 2020-2021 Science Olympiad Protein Modeling Event Webspace
This year’s event focuses on the CRISPR bacterial adaptive immunity system and base editing using cytidine deaminase. Explore the introduction video and additional content below to prepare for this year's event.
Introduction Video
How to Prepare for this Year's Event
The HHMI BioInteractive CRISPR Website
This is an excellent resource for understanding the basics of the CRISPR Cas9 protein. The first section of the website walks through the structure and function of Cas9, and the second section is a series of videos from leading researchers in the CRISPR field.
Expanding on the HHMI Resources
Below are a series of CBM videos to expand on the HHMI resources and further explore the CRISPR System and base editing with the Cas9 protein.
audio 3The CRISPR Adaptive Immunity System
Where Did CRISPR Come From?
The Cas9 Schematic Model
Additional Context to the CRISPR Adaptive Immunity System
Additional Videos to Help Get Your Head
Around CRISPR. . . and Base Editing
CRISPR technology has already revolutionized the molecular biosciences. Many excellent videos have appeared that explain how this technology works. Here are a few of our favorites – directed toward a variety of audiences and presented at varying levels of detail.
What Is CRISPR?
Bozeman Science / Paul Andersen provides a very nice overview of the CRISPR System.
CRISPR Gene Editing and Beyond
A protein-centric video of CRISPR, from Nature (a weekly science journal).
CRISPR: History of Discovery
A Singapore NIE video that documents the historical milestones in the discovery of the CRISPR system.
Ask the CBM!
Answered Questions:
Additional answers will be added as more questions are submitted and audio recordings are completed.
1. How can I model cas9 and cytidine deaminase in one Jmol File?
2. I'm looking for the articles Dr. Herman discussed in his videos?
Link to "This Year's Theme" webpage, which has both papers available to download in "Tab 3: The 2020-21 Prebuild Protein"
3. What is the difference between crRNA and tracrRNA?
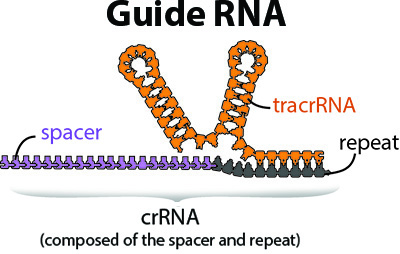
4. Which Molecule of the Month article is appropriate for this year's event?
Link to the RCSB Protein Data Bank's Molecule of the Month on CRISPR
5. What information on the PDB file, not the data visualization environment, is important?
Link to the video describing the RCSB Protein Databank and how to prepare for the onsite "Jmol Exploration".
6. Why are there two papers, and are they both necessary?
7. In the pre-build visualization environment there seems to be some missing, because it goes from 241 to 250. What happened to the 8 amino acids in between?
8. How do we know which sidechains are important and relevant to the competition model? What types of structures can be of significance to add to the pre-build model?
9. How many “cheat sheets” are you allowed to bring into the competition?
10. How do you figure out what sidechains an amino acid is interacting with? And once you determine that, is there any way to know what type of interaction it is making?
11. What does cytidine deaminase do?
12. What resources do you recommend for the new onsite Jmol Exploration portion of the competition?
13. The Schiffer Paper mentions a Poly T Background. What does this mean?
14. Where on the RCSB Protein Databank are we supposed to find 3D coordinates for amino acids?
15.What does the purple mean when you color by “structure” in Jmol?
16. Why are negatively charged sidechains called “acidic”?
17. Do the cysteine amino acids form disulfide salt bridges?
18. We have noticed that the cytidine deaminase structure includes a short piece of DNA. Do we need to know anything about this?
19. The amino acid E72 is described in the paper as a glutamic acid, but in the PDB file structure it appears to be an alanine. Why is that?
20. What is the “schematic fusion protein model”?
21. How do you view the endcaps of the protein in Jmol?
22. How can I find out the function of relevant sidechains on the prebuild model?
23. Why is the pyrimidine tyrosine switched to uracil when converting from DNA to RNA while the other nucleotides stay constant?
24. The Prebuild PDB file shows three ions attached to the protein. Are these important? If so, what do they do?
25. It seems like histidine 29 is an incredibly important sidechain, but it isn’t within the range of the Prebuild protein model. Why is that?
26. Should we include important sidechains where the single stranded DNA interacts with the protein? If not, what are some other things that can set us aside from other teams?
27. How do we know that the CRISPR Cas9 with the cytidine deaminase fusion protein does not have off-target effects, and why is this the case?
28. Will all the questions about CRISPR come from the CBM website and articles, or should we use outside information to prepare?
29. On the rules, under 4b3, it says we should display the larger fusion protein of the BE4 expression plasmid. Are we expected to make a separate model of this, or should we incorporate it into our cytidine deaminase model?
30. The national rubric links to a plasmid on Addgene that contains APOBEC1. Since the pre-build model is APOBEC3a, what should we do to incorporate the rest of the fusion protein with the APOBEC3a. Should we assume that they are close enough?
31. During the competition, are we allowed to access the RCSB Protein Databank?
32. What is the process of long-patch BER, or basic excision repair?
33. Why does the Cas9 system need to save a DNA sequence from a bacteriophage, if it can just cut all viral DNA that enters the bacteria regardless of if the bacteria has encountered it before and has a part of its sequence?
36. Is the section on the different interactions between A3A and RA3G.ntd significant for the event at all?
34. What do the different colored highlights mean on the papers?
35. Do we have to explain the purpose behind every add-on to our pre-build model?
36. Do we have to know the history of CRISPR and do we have to know protein folding principles?
37. Is there a specific functional reason for why we are only modeling amino acids residues 31-145? Why were the beginning and end amino acids excluded?
38. How should I determine which sequence would most likely be a PAM domain or PAM binding site?
In order to truly appreciate and successfully model this year's Protein Modeling Event structures, a thorough understanding of protein structure is needed. This section will explore these amazing macromolecules in more detail using suggested physical model kits, online resources and additional websites.
Protein Structure
Proteins are long linear sequences of amino acids
that fold into complex 3-dimensional shapes
following basic principles of chemsitry.
In this first of two videos on protein folding, Tim Herman, Ph.D. from the Center for BioMolecular Modeling uses the Water Cup from 3D Molecular Designs to demonstrate some basic principles of chemistry.
In this second of two videos on protein folding, Tim Herman, Ph.D. from the Center for BioMolecular Modeling uses the Amino Acid Starter Kit from 3D Molecular Designs to demonstrate how the basic principles of chemistry directly affect protein folding.
In this video, Tim Herman, Ph.D. from the Center for BioMolecular Modeling uses the Alpha-helix Beta-sheet Construction Kit from 3D Molecular Designs to demonstrate the form and function of secondary structures in proteins.

Protein Databank (www.rcsb.org) Resources
Learn about protein structure and function with this overview printout and video developed by the RCSB Protein Databank.
- What is a Protein? (PDF)
- What is a Protein? (video)

3D Molecular Designs Educational Kits
These engaging, hands-on kits make learning protein structure basics easy. Users will fold a protein while exploring how the chemical properties of amino acids determine its final structure.
DNA Structure
DNA carries your genetic information in that it determines the sequence of amino acids in your proteins. These protein sequences, in turn, determine the shape and structure of your proteins. Proteins then work like nano-scale molecular machines, carrying out countless different tasks in your body.
The human genome is divided into 23 chromosomes pairs, found in the nucleus of every cell. To keep these long linear polymers of DNA from getting all tangled up, the DNA of each chromosome is packaged into repeating structural units called nucleosomes. The DNA exists as a double–stranded structure with two twisting backbones running in opposite directions and four different bases: adenosine (A), thymine (T), guanine (G), and cytosine (C).
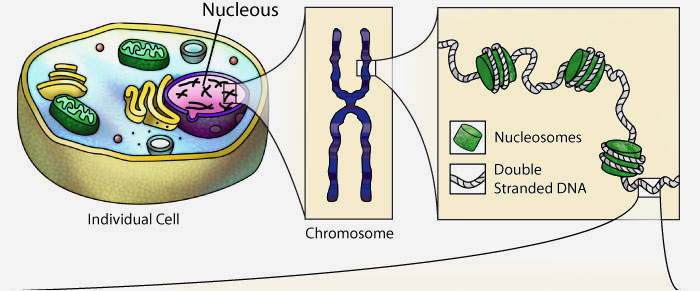
The visual representation of DNA above is completely interactive and can be rotated in 3-dimensions by clicking and dragging with your mouse.
The four types of bases that make up DNA can form base-pairs between the two strands of double-stranded DNA. Adenosine only pairs with thymine and guanine only pairs with cytosine, making the two strands of DNA complimentary. The sequence of bases is often represented with abbreviated letters as shown below. It is this order of DNA bases that contains the key information needed for creating proteins and passing along genetic information.
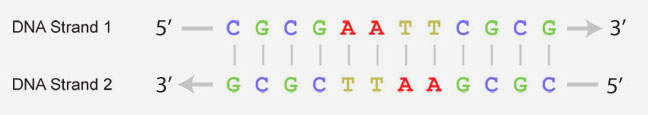
For more information on DNA structure, explore the RCSB PDB's Molecule of the Month feature on DNA article by the molecular illustrator, David Goodsell.
APOBEC3A Cytidine Deaminase Protein
Based on 5keg.pdb

 NOTE: This is a different structure/sequence than originally stated
NOTE: This is a different structure/sequence than originally stated
in the Science Olympiad rules. The correct structure/sequence is
amino acids 31-145 of chain A of 5keg.pdb

Below are the papers (i.e., the primary citations) described in the video above and referencing the structure to be modeled for this year's prebuild.
Crystal structure of APOBEC3A bound to single-stranded DNA reveals structural basis for cytidine deamination and specificity
Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage
The pre-build model will be created using a purchased "pre-build" Mini-Toober kit from www.3dmoleculardesigns.com, or with found materials of the participants' choosing such as KwikTwist tie-down ropes.
The same pre-build model will be used at each level of the competition (invitational, regional, state and national). Participants will take their pre-build model home after each event.
The pre-build model must be impounded prior to the beginning of the event (usually the morning of the event. The pre-build model should be submitted with a 4"x6" card with a written description of the model's colors and creative additions, addressing what, how, and why. Check the official rules for details
View the 2020-2021 PreBuild Design Environment
Exploring the RCSB Protein Databank for the Onsite Jmol Exploration
This year, instead of modeling a protein during the on-site competition, teams will answer questions as part of a "Jmol Exploration".
The purpose of the Jmol exploration component of this year’s event is to allow students to demonstrate their skill in using Jmol to manipulate and explore a pdb file, and navigate the RCSB Protein Data Bank website (www.rcsb.org).
The video below will describe the RCSB Protein Databank Website, and the features teams should be familiar with for the onsite "Jmol Exploration" portion of the competition.
